Many large data sets, such as gigapixel images, geographic maps, or networks, require exploration of annotations at different levels of detail. We call an annotation a region in the visualization that contains some visual patterns (called annotated pattern henceforth). These annotated patterns can either be generated by users or derived computationally. However, annotated patterns are often magnitudes smaller than the overview and too small to be visible. This makes tasks such as exploration, searching, comparing, or contextualizing challenging, as considerable navigation is needed to overcome the lack of overview or detail.
Exploring annotated patterns in context is often needed to assess the relevance of patterns and to dissect important from unimportant regions. For example, computational biologists study thousands of small patterns in large genome interaction matrices [1] to understand which physical interactions between regions on the genome are the driving factor that defines the structure of the genome. In astronomy, researchers are exploring and comparing multiple heterogeneous galaxies and stars within super high-resolution imagery [2]. In either case, inspecting every potentially important region in detail is simply not feasible.

Exploring visual details of these annotated patterns in multiscale visualizations requires a tradeoff between several conflicting criteria (Fig. 1). Patterns must be visible for inspection and comparison (detail). Enough of the overview needs to be visible to provide context for the patterns (context). And the detailed pattern representations must be close to its actual position in the overview (locality).
Current interactive navigation and visualization approaches, such as focus+context, overview+detail, or general highlighting techniques, address some but not all of these criteria and become difficult as repeated viewport changes, multiple manual lenses, or separate views at different zoom levels are required, which stress the user’s mental capacities.
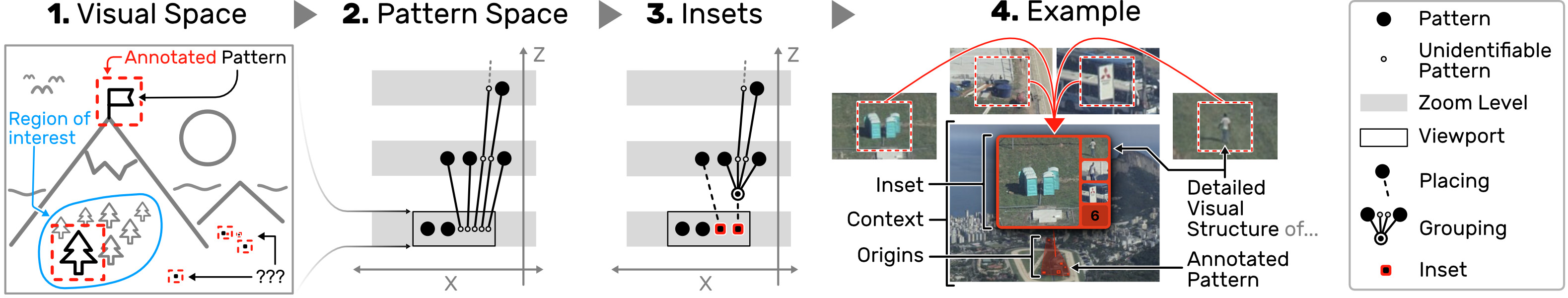
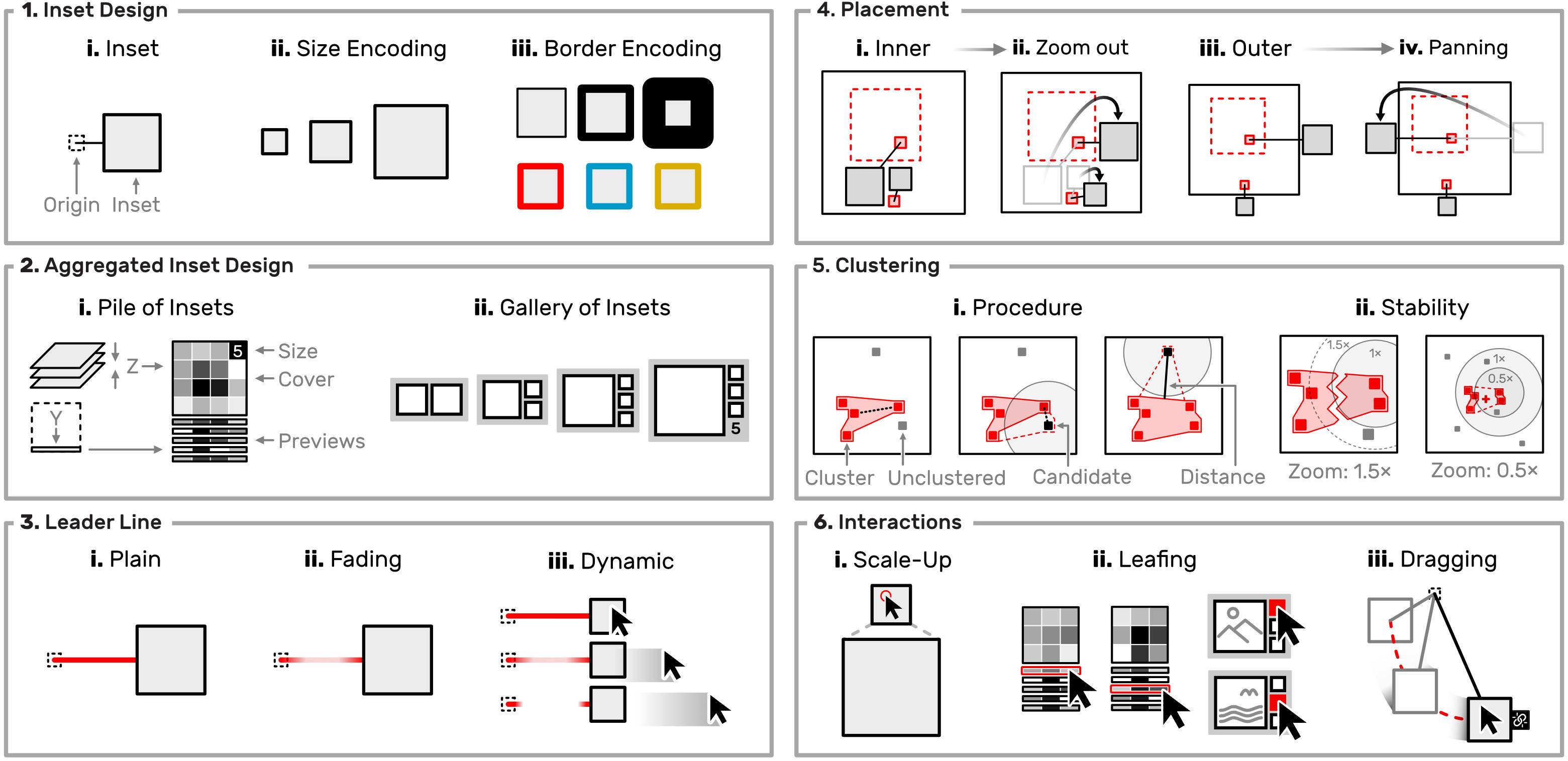
Scalable Insets is a scalable visualization technique for interactively exploring and navigating large numbers of annotated patterns in multiscale visualizations. Scalable Insets support users in early exploration through multifocus guidance by dynamically placing magnified thumbnails of annotated patterns as insets within the viewport (Fig. 2). The entire design (Fig. 3) is focused on scalability. To keep the number of insets stable, we developed dynamic grouping to cluster patterns based on their location, type, and the user’s viewport.
The degree of clustering constitutes a tradeoff between context and detail. Groups of patterns are visually represented as a single aggregated inset to accommodate for detail. Details of aggregated patterns are gradually resolved as the user navigates into certain regions. We also present two dynamic mechanisms for placing insets either within the overview or on the overview’s boundaryto allow flexible adjustment to locality. With Scalable Insets, the user can rapidly search, compare, and contextualize large pattern spaces in multiscale visualizations.
We implemented Scalable Insets as an extension to HiGlass [3], a flexible web application for viewing large tile-based datasets. The implementation currently supports gigapixel images, geographic maps, and genome interaction matrices. In a qualitative user study six computational biologists explored features in a genome interaction matrices using Scalable Insets.
Their feedback shows that our technique is easy to learn and effective in biological data exploration. Results of a controlled user study with 18 novice users comparing both placement mechanisms of Scalable Insets to a standard highlighting technique show that Scalable Insets reduced the time to find annotated patterns by up to 45% and improved the accuracy in comparing pattern types by up to 32 percentage points.